Abstract
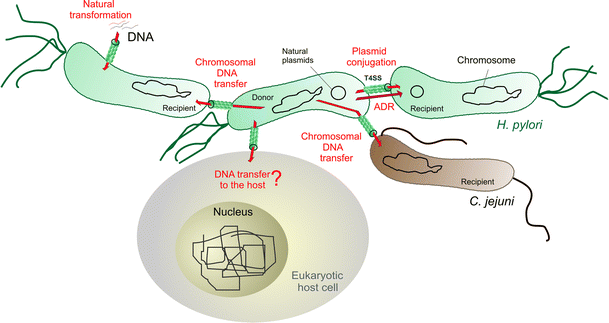
Genetic recombination may be evolutionarily important in speeding the adaptation of organisms to new environments and in eliminating deleterious mutations. Here, we describe polymerase chain reaction (PCR), hybridization, and DNA sequence-based evidence of six such exchanges between two Helicobacter pylori strains during natural mixed infection of a patient in Lithuania. One parent strain contained the 37 kb long virulence-associated cag pathogenicity island (PAI), and the other strain lacked this PAI.
Most of the patient’s H. pylori recombinants had descended from the cag+ parent but had been converted to cag- during infection. This had resulted from the transfer of DNA containing the ’empty site’ allele from the cag strain and homologous recombination, not excision of PAI cag without DNA transfer. Other cases of recombination involved genes for an outer membrane protein (omp5 and omp29; also called HP0227 and HP1342) and a putative phosphoenolpyruvate synthase (ppsA; HP0121).
Replacement of a short patch of DNA sequence (36-124 bp) was also observed. Since the chance of forming any given recombinant is small, the abundance of recombinants in this patient suggests selection for particular recombinant genotypes during years of chronic infection. We suggest that genetic exchange between unrelated H. pylori strains, as documented here, is important due to the diversity of this gastric pathogen and its human hosts. Certain H. pylori recombinants may grow better in a given host than either parent. Growth vigour, in turn, could affect the severity of the disease that infection can cause.
Materials And Methods
- Materials
A well-characterized strain, H. pylori, was provided by the Department of Microbiology at Chongqing University of Medical Sciences. Top10 strains, BL21 E. coli and plasmid pET32a(+) were submitted by the Institute of Viral Hepatitis, Chongqing University of Medical Sciences. Restriction enzymes (Hind III, BamH I) and T4 DNA ligase were purchased from Promega, Tag DNA polymerase was produced by the Department of Immunology of Old Beijing Medical University. Isopropyl-β-D-thiogalactopyranoside (IPTG) primers, dNTPs and oligonucleotides were obtained from Sigma Chemical Co., etc.
- Cloning of the OMP gene from Hp Mr26000
Oligonucleotide primers were designed to amplify the H. pylori open reading frame (ORF) of the outer membrane protein Mr26000 based on the published genome sequence. The primers were designed with an incorporated BamH I site at the 5′ end and a Hind III site at the 3′ end as follows (5′-3′): GCGGATCCATGTTAGTTACAAAACTTGCC (forward) and AAGCTTAATGGAATTTTCTTT (backwards). Genomic DNA prepared from Chongqing H. pylori strains was used as a template in the PCR.
The PCR cycle consisted of 30 cycles of denaturation at 94 °C for 60 s, annealing at 58 °C for 45 s, with an extension step at 72 °C for 90 s. The products were visualized on a 10 g·L⁻¹ agarose gel and purified using a PCR purification kit. After digestion with restriction enzymes BamH I and Hind III simultaneously, purified products were cloned into compatible sites of pET32a(+) expression vectors using T4 DNA ligase in a 4∶1 molar ratio at 4 °C. overnight.
Fifty μL of Top10 incubated at 37 °C overnight was added to 2 mL of Luria-Bertani broth and routinely cultured at 37 °C and shaken at 300 r·min-1 for 4 h. When the optical density at 600 nm was 0.5, it was ultracentrifuged at 10,000 r·min-1 at room temperature (RT) for 2 min. The resulting pools were suspended with 100 mmol·L⁻¹ 150 µL CaCL2 and incubated at 0 °C for 2 h.
Ten μL of connected products (top) were resuspended and incubated at 0 °C for 30 min, 42 °C for 2 min, and 0 °C for 2 min, respectively. Finally, it was incubated at 37 °C at 180 r min-1 for 30 min after adding 1 mL of LB broth, 200 μL were collected and spread on an LB plate containing 100 mg•L⁻¹ ampicillin as selectable broth. marker and incubated at 37 °C overnight.
- Extraction and expression of recombinant plasmid
The next day, the single cloned bacterial droplet was selected and cultured in 2 mL of LB broth containing 100 mg•L⁻¹ ampicillin at 37 °C overnight at 300 r•min-1, then plasmids were extracted. recombinants and examined with plasmid extraction. The kits according to the manufacturer’s instructions, meanwhile, identified by PCR and restriction enzyme digestion.
Recombinant plasmids were selected and transformed into competent E. coli BL21(DE3) strains using standard procedures. E. coli BL21 strains to contain recombinant plasmid were grown to mid-log phase (optical density at 600 nm = 0.5 to 1.0) and expression of the fusion proteins was induced by the addition of 0.5- 4.0 mmol·L⁻¹ of IPTG for 4 h. . After induction, the bacteria were harvested by ultracentrifugation at 12,000 r·min-1, resuspended in protein buffer and boiled for 5 min. Total protein was subjected to SDS-PAGE gel electrophoresis and coomassie stained.
- Western blot analysis
Briefly, the Mr26000 OMP was purified using Ni-NTA agarose resin after the bacteria were grown and decomposed in the microwave at 600 W × 35% power for 40 min, ultracentrifuged (10000 g, 15 min, 4 °C). C) and then quantified. H. pylori outer membrane protein-specific antibody Mr26000 was produced after subcutaneous immunization of New Zealand rabbits, while age-matched control rabbits were immunized with PBS as previously described.
Serum antibody specificity was determined by ELISA or immunoblotting after electrophoretic transfer of (150 g·L⁻¹ acrylamide)H separated by SDS-PAGE. pylori Mr26000 PVDF membrane outer membrane protein of 0.45 μm pore size. After a 30 min wash in Tris-saline blotting buffer, antigen-impregnated PVDF strips were incubated with the rabbit sera for 2 h at RT. After washing, bound rabbit antibodies were detected by incubating the strips in alkaline phosphatase-conjugated goat anti-rabbit IgG antibody for 1 hour at room temperature.
- Prophylactic immunization
Six to eight-week-old mice were immunized three times by subcutaneous immunization using Mr26000 OMP emulsified with Freund’s adjuvant at intervals of 1, 14 and 21 days, respectively, to produce antibodies responsive to the outer membrane protein Mr26000. The dose consisted of 1 mL (100 mg·L⁻¹) of purified Mr26000 OMP and 1 mL of complete Freund’s adjuvant. Subsequently, the dose consisted of 0.5 mL of OMP and 0.5 ml of Freund’s adjuvant in complement. Age-matched control mice were immunized with PBS. Antibody titers in immunized mice were monitored by ELISA with the purified fusion protein.
Mice were challenged with a single dose of 108 H. pylori organisms 7 days after the last immunization. Twenty-eight days after the challenge, mice were sacrificed by cervical dislocation. The stomach was removed from each animal, sectioned in two longitudinally and pinned apart. Full-thickness tissue was taken from the antrum-body area of one half of each stomach and placed in 0.2 ml of urease test medium.
Urease activity in the sample, identified by a distinctive colour change in the medium, was assessed after 24 h incubation at room temperature. The rest of the stomach was fixed in 100 mL·L⁻¹ buffered formalin and embedded in paraffin. Longitudinal sections, stained with a modified May-Grunwald Giemsa stain, were scanned in full length under light microscopy. Mice were considered protected or not according to the previous report.
- Statistic analysis
The student’s t-test was used to assess the presence or absence of experimental infection in test and control animals, as well as the anti-Mr26000 outer membrane protein response to immunization. Values of p < 0.05 were considered statistically significant.
Conclusion:
Mr26000 OMP may be a candidate vaccine to prevent H. pylori infection.